Language Details¶
The following is a fairly extensive introduction to the grama language. This
is (admittedly) not a great place to start learning how to use py_grama, but
is instead provided as a reference.
grama is a conceptual language, and py_grama is a code implementation of
that concept. This page is a description of both the concept and its
implementation.
Running example¶
We’ll use a running example throughout this page; the built-in py_grama Cantilever Beam model.
import grama as gr
from grama.models import make_cantilever_beam
md_beam = make_cantilever_beam()
md_beam.printpretty()
model: Cantilever Beam
inputs:
var_det:
w: [2, 4]
t: [2, 4]
var_rand:
H: (+1) norm, {'loc': 500.0, 'scale': 100.0}
V: (+1) norm, {'loc': 1000.0, 'scale': 100.0}
E: (+0) norm, {'loc': 29000000.0, 'scale': 1450000.0}
Y: (-1) norm, {'loc': 40000.0, 'scale': 2000.0}
functions:
cross-sectional area: ['w', 't'] -> ['c_area']
limit state: stress: ['w', 't', 'H', 'V', 'E', 'Y'] -> ['g_stress']
limit state: displacement: ['w', 't', 'H', 'V', 'E', 'Y'] -> ['g_disp']
Objects¶
grama focuses on two categories of objects:
- data (
df): observations on various quantities, implemented by the Python package Pandas - models (
md): functions and a complete description of their inputs, implemented by py_grama
For readability, we suggest using prefixes df_ and md_ when naming DataFrames and models.
Data¶
Data are observations on some quantities. Data often come from the real world,
but data are also used to inform models, and models can be used to generate new
data. py_grama uses the Pandas DataFrame implementation to represent data.
Since data operations are already well-handled by Pandas, py_grama uses the
existing Pandas infrastructure and focuses on providing tools to handle models
and their interface with data.
Models¶
Models in grama have both functions and inputs. py_grama implements models
in the Model class, which in turn have three primary objects:
Domain: Defines bounds on variablesDensity: Defines a joint density for variables- List of
Functions: Maps variables to outputs
Domain¶
The domain of the model defines bounds for all the variables. If a variable is
not included in the domain object, it is assumed to be unbounded. The model
above has bounds on t, w, both of which are [2, 4].
Density¶
The density of the model defines a joint density for the random variables. If
a variable is included in the density it is random, otherwise it is
deterministic. The model above has a joint density on H, V, E, Y.
The model summary gives details on each marginal distribution.
Functions¶
A function has a set of variables, which map to a set of outputs; for instance,
the cross-sectional area function above maps ['w', 't'] -> ['c_area']. The
other functions take more variables, all of which map to their respective
outputs.
Inputs¶
The full set of model inputs are organized into:
| Deterministic | Random | |
|---|---|---|
| Variables | md.var_det |
md.var_rand |
| Parameters | md.density.marginals[var].d_param |
(Future*) |
- Variables are inputs to the model’s functions
- Deterministic variables are chosen by the user; the model above has
w, t - Random variables are not controlled; the model above has
H, V, E, Y
- Deterministic variables are chosen by the user; the model above has
- Parameters are inputs to the model’s density
- Deterministic parameters are currently implemented; these are listed under
var_randwith their associated random variable - Random parameters* are not yet implemented
- Deterministic parameters are currently implemented; these are listed under
The full set of variables is determined by the domain, density, and functions.
Formally, the full set of variables is given (in pseudocode) by domain.var + [f.var for f in functions]. The set of random variables is then given by
domain.var + [f.var for f in functions] - density.marginals.keys(), while the
deterministic variables are the remainder var_det = var_full - var_rand.
Verbs¶
Verbs are used to take action on different grama objects. We use verbs to generate data from models, build new models from data, and ultimately make sense of the two.
The following table summarizes the categories of py_grama verbs. Verbs take
either data (df) or a model (md), and may return either object type. The
prefix of a verb immediately tells one both the input and output types. The
short prefix is used to denote the pipe-enabled version of a verb.
| Verb Type | Prefix (Short) | In | Out |
|---|---|---|---|
| Evaluate | eval_ (ev_) |
md |
df |
| Fit | fit_ (ft_) |
df |
md |
| Transform | tran_ (tf_) |
df |
df |
| Compose | comp_ (cp_) |
md |
md |
| Plot | plot_ (pt_) |
df |
(Plot) |
Since py_grama is focused on models, the majority of functions lie in the
Evaluate, Fit, and Compose categories, with only a few original Transform
utilities provided. Some shortcut plotting utilities are also provided
for covenience.
py_grama verbs are used to both build and analyze models.
Model Building¶
The recommended way to build py_grama models is with composition calls.
Calling Model() creates an “empty” model, to which one can add.
md = gr.Model()
md.printpretty()
model: None
inputs:
var_det:
var_rand:
functions:
We can then use Compose functions to build up a complete model step-by step. We recommend starting with the functions, as those highlight the required variables.
md = gr.Model("Test") >> \
gr.cp_function(
fun=lambda x: [x[0], x[1]],
var=["x0", "x1"],
out=2,
name="Identity"
)
md.printpretty()
model: Test
inputs:
var_det:
x0: (unbounded)
x1: (unbounded)
var_rand:
functions:
Identity: ['x0', 'x1'] -> ['y0', 'y1']
Note that by default all of the variables are assumed to be deterministic. We can override this by adding marginal distributions for one or more of the variables.
md = gr.Model("Test") >> \
gr.cp_function(
fun=lambda x: [x[0], x[1]],
var=["x0", "x1"],
out=2,
name="Identity"
) >> \
gr.cp_marginals(
x1=dict(dist="norm", loc=0, scale=1)
)
md.printpretty()
model: Test
inputs:
var_det:
x0: (unbounded)
var_rand:
x1: (+0) norm, {'loc': 0, 'scale': 1}
functions:
Identity: ['x0', 'x1'] -> ['y0', 'y1']
The marginals are implemented in terms of the Scipy continuous
distributions; see the
variable gr.valid_dist.keys() for a list of implemented marginals. When
calling gr.comp_marginals(), we provide the target variable name as a keyword
argument, and the marginal information via dictionary. The marginal shape is
specified with the “dist” keyword; all distributions require the loc, scale
parameters, but some require additional keywords. See gr.param_dist for a
dictionary mapping between distributions and parameters.
Once we have constructed our model, we can analyze it with a number of tools.
Model Analysis¶
One question in model analysis is to what degree the random variables affect the
outputs. A way to quantify this is with Sobol’ indices (Sobol’, 1999). We
can estimate Sobol’ indices in py_grama with the following code.
df_sobol = \
md_beam >> \
gr.ev_hybrid(n=1e3, df_det="nom", seed=101) >> \
gr.tf_sobol()
print(df_sobol)
eval_hybrid() is rounding n...
w t c_area g_stress g_disp ind
0 NaN NaN NaN -0.03 0.28 S_E
0 NaN NaN NaN 0.35 0.21 S_H
0 NaN NaN NaN 0.33 0.64 S_V
0 NaN NaN NaN 0.31 0.02 S_Y
0 0.0 0.0 0.0 -345263.39 0.01 T_E
0 0.0 0.0 0.0 4867712.30 0.01 T_H
0 0.0 0.0 0.0 4577175.85 0.02 T_V
0 0.0 0.0 0.0 4224965.01 0.00 T_Y
0 0.0 0.0 0.0 13758547.04 0.04 var
The normalized Sobol’ indices are reported with S_[var] labels; they indicate
that g_stress is affected roughly equally by the inputs H,V,Y, while
g_disp is affected about twice as much by V as by E or H. Note that the
Sobol’ indices are only defined for the random variables—since Sobol’
indices are defined in terms of fractional variances, they are only formally
valid for quantifying contributions from sources of randomness.
Under the hood gr.eval_hybrid() attaches metadata to its resulting DataFrame,
which gr.tran_sobol() detects and uses in post-processing the data.
py_grama also provides tools for constructing visual summaries of models. We
can construct a sinew plot with a couple lines of code. First we inspect the
design:
md_beam >> \
gr.ev_sinews(n_density=50, n_sweeps=10, df_det="nom", skip=True) >> \
gr.pt_auto()
 beam sinew results
beam sinew results
The “sinews” are sweeps across random variable space which start at random locations, and continue parallel to the variable axes. Evaluating these samples allows us to construct a sinew plot:
md_beam >> \
gr.ev_sinews(n_density=50, n_sweeps=10, df_det="nom", skip=False) >> \
gr.pt_auto()
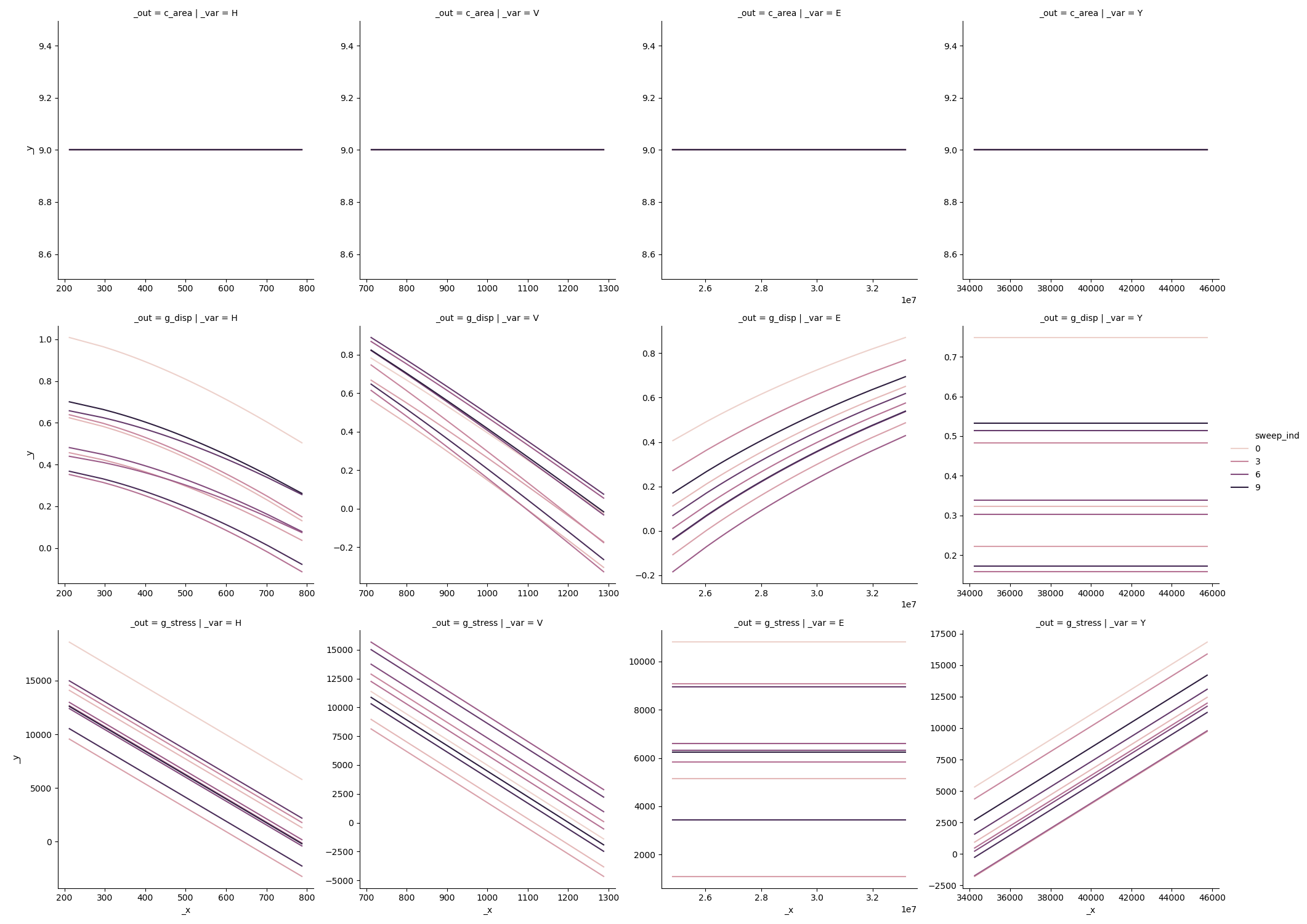 beam sinew results
beam sinew results
Here we can see that inputs H,E tend to saturate in their effects on g_disp,
while V is linear over its domain. This may explain the difference in
contributed variance seen above via Sobol’ indices.
By providing tools to quickly perform different analyses, one can quickly get a
sense of model behavior using py_grama.
Layers and Defaults¶
py_grama is built around layers and defaults. As much as is possible
py_grama is designed to provide sensible defaults “out-of-the-box”. We saw the
concept of layers above in the model building example. The following example
shows defaults in action.
Example Defaults: gr.eval_monte_carlo()¶
Attempting to provide no arguments to gr.eval_monte_carlo() yields the
following error:
df_res = md_beam >> gr.ev_monte_carlo()
...
ValueError: df_det must be DataFrame or 'nom'
One can sample over the random variables given their joint density, but this
tells us nothing about how to treat the deterministic variables. The error
message above tells us that we have to define the deterministic variable levels
through df_det. To perform simple studies, we can explicitly limit attention
to the nominal conditions.
df_res = md_beam >> gr.ev_monte_carlo(df_det="nom")
print(df_res.describe())
H V E ... c_area g_stress g_disp
count 1.000000 1.000000 1.000000e+00 ... 1.0 1.00000 1.000000
mean 505.743982 906.946892 2.824652e+07 ... 9.0 9758.06826 0.438044
std NaN NaN NaN ... NaN NaN NaN
min 505.743982 906.946892 2.824652e+07 ... 9.0 9758.06826 0.438044
25% 505.743982 906.946892 2.824652e+07 ... 9.0 9758.06826 0.438044
50% 505.743982 906.946892 2.824652e+07 ... 9.0 9758.06826 0.438044
75% 505.743982 906.946892 2.824652e+07 ... 9.0 9758.06826 0.438044
max 505.743982 906.946892 2.824652e+07 ... 9.0 9758.06826 0.438044
[8 rows x 9 columns]
By default gr.eval_monte_carlo() will draw a single sample; this leads to the
NaN standard deviation (std) results. We can override this default behavior
by providing the n keyword.
df_res = md_beam >> gr.ev_monte_carlo(df_det="nom", n=1e3)
print(df_res.describe())
eval_monte_carlo() is rounding n...
H V E ... c_area g_stress g_disp
count 1000.000000 1000.000000 1.000000e+03 ... 1000.0 1000.000000 1000.000000
mean 499.776481 1001.908814 2.899238e+07 ... 9.0 6589.731053 0.333387
std 100.878972 102.929434 1.523033e+06 ... 0.0 3807.292688 0.199576
min 168.466610 635.220875 2.427615e+07 ... 9.0 -5830.363269 -0.339594
25% 433.466349 933.698938 2.799035e+07 ... 9.0 3921.581411 0.200657
50% 500.201964 1002.514072 2.896188e+07 ... 9.0 6633.083925 0.339702
75% 569.159912 1065.489928 2.993924e+07 ... 9.0 9161.066426 0.465231
max 798.543698 1327.618914 3.378264e+07 ... 9.0 18682.531892 1.045742
[8 rows x 9 columns]
Formally 1e3 is a float, which is not a valid iteration count. The routine
gr.eval_monte_carlo() informs us that it first rounds the given value before
proceeding. Here we can see some variation in the inputs and outputs, though
c_area is clearly unaffected by the randomness.
We can also provide an explicit DataFrame to the df_det argument. The
gr.eval_monte_carlo() routine will automatically take an outer product of
the deterministic settings with the random samples; this will lead to a
multiplication in sample size of df_det.shape[0] * n. Since this can get quite
large, we should reduce n before proceeding. We can also delay evaluation
first with the skip keyword, and inspect the design first before evaluating
it.
df_det = pd.DataFrame(dict(
w=[3] * 10,
t=[2.5 + i/10 for i in range(10)]
))
df_design = md_beam >> gr.ev_monte_carlo(df_det=df_det, n=1e2, skip=True)
print(df_design.describe())
eval_monte_carlo() is rounding n...
H V E Y w t
count 1000.000000 1000.000000 1.000000e+03 1000.000000 1000.0 1000.000000
mean 485.021539 989.376268 2.890355e+07 40135.172902 3.0 2.950000
std 112.546373 92.381768 1.462286e+06 2135.788506 0.0 0.287372
min 137.606474 764.764234 2.586955e+07 33320.789307 3.0 2.500000
25% 411.652958 927.448139 2.763002e+07 39104.255172 3.0 2.700000
50% 490.765903 1002.513232 2.903287e+07 40121.853496 3.0 2.950000
75% 561.317283 1046.038620 2.994359e+07 41279.345157 3.0 3.200000
max 726.351812 1239.473097 3.176696e+07 45753.627265 3.0 3.400000
If we are happy with the design (possibly after visual inspection), we can pass the input DataFrame to the straight evaluation routine
df_res = md_beam >> gr.ev_df(df=df_design)
print(df_res.describe())
H V E ... c_area g_stress g_
disp
count 1000.000000 1000.000000 1.000000e+03 ... 1000.000000 1000.000000 1000.00
0000
mean 510.457048 1003.972079 2.887534e+07 ... 8.850000 4555.508097 0.12
0805
std 105.566743 91.443354 1.486829e+06 ... 0.862116 7027.137049 0.59
9538
min 269.102628 830.234813 2.553299e+07 ... 7.500000 -18370.732185 -1.84
8058
25% 437.342719 939.490354 2.794973e+07 ... 8.100000 -385.620306 -0.29
7274
50% 515.387982 995.468715 2.868415e+07 ... 8.850000 5347.337598 0.23
2133
75% 584.899669 1054.815972 2.982724e+07 ... 9.600000 9824.615141 0.60
9096
max 775.221819 1257.095025 3.369872e+07 ... 10.200000 21238.954122 1.23
6387
[8 rows x 9 columns]
Functional Programming (Pipes)¶
Functional programming
touches both the practical and conceptual aspects of the language. py_grama
provides tools to use functional programming patterns. Short-stem versions of
py_grama functions are pipe-enabled, meaning they can be used in functional
programming form with the pipe operator >>. These pipe-enabled functions are
simply aliases for the base functions, as demonstrated below:
df_base = gr.eval_nominal(md_beam, df_det="nom")
df_functional = md_beam >> gr.ev_nominal(df_det="nom")
df_base.equals(df_functional)
True
Functional patterns enable chaining multiple commands, as demonstrated in the following Sobol’ index analysis. In nested form using base functions, this would be:
df_sobol = gr.tran_sobol(gr.eval_hybrid(md_beam, n=1e3, df_det="nom", seed=101))
From the code above, it is difficult to see that we first consider md_beam, perform a hybrid-point evaluation, then use those data to estimate Sobol’ indices. With more chained functions, this only becomes more difficult. One could make the code significantly more readable by introducing intermediate variables:
df_samples = gr.eval_hybrid(md_beam, n=1e3, df_det="nom", seed=101)
df_sobol = gr.tran_sobol(df_samples)
Conceptually, using pipe-enabled functions allows one to skip assigning intermediate variables, and instead pass results along to the next function. The pipe operator >> inserts the results of one function as the first argument of the next function. A pipe-enabled version of the code above would be:
df_sobol = \
md_beam >> \
gr.ev_hybrid(n=1e3, df_det="nom", seed=101) >> \
gr.tf_sobol()
References¶
- I.M. Sobol’, “Sensitivity Estimates for Nonlinear Mathematical Models” (1999) MMCE, Vol 1.